ACRES - Software - Matlab Parallel
- 1Overview
- 2Run Interactively from your PC
- 3Cluster Profile Configuration and Validation
- 4Running an Interactive Parallel Session
- 5Submitting a Batch Job to the Cluster and Retrieving Results
- 6Custom queue submission parameters
Overview
This guide describes how to run Matlab Parallel scripts, using ACRES resources. It is assumed that you already have a Matlab script that is able to run in parallel, using Matlab Parallel Computing Toolbox. For assistance creating such a script, please refer to the Matlab documentation.
Run Interactively from your PC
To use the Parallel Computing Toolbox (from here on: 'PCT') with ACRES, you must configure your local Matlab client and allow ACRES to communicate with it through your Firewall.
This page aims to provide a template of the configuration necessary to make this work.
Please be aware that you must already have a license for the PCT, and already have it installed appropriately before proceeding on this page, and please note that only the supported versions of MATLAB, listed below, will work.
Version Matters!
Make sure you are using one of the supported versions of Matlab. Only the supported versions will work.
Troubleshooting Guide Available
If you encounter any errors, please check the Troubleshooting section on this page for possible solutions.
Supported Versions
R2022a or newer
Step 1. Set Firewall Rule
You need to set up a firewall rule to allow ACRES and MATLAB to freely communicate, which means setting up a rule to accept signals from the ACRES head node (ip address: 128.153.5.166) to a certain range of ports that MATLAB is configured to use on your local machine (by default these ports are 27370-27470). The process of setting up this rule is done differently depending on your local Operating System and firewall software. Some step-by-step guides are given below for some common Operating Systems and software.
Windows 10
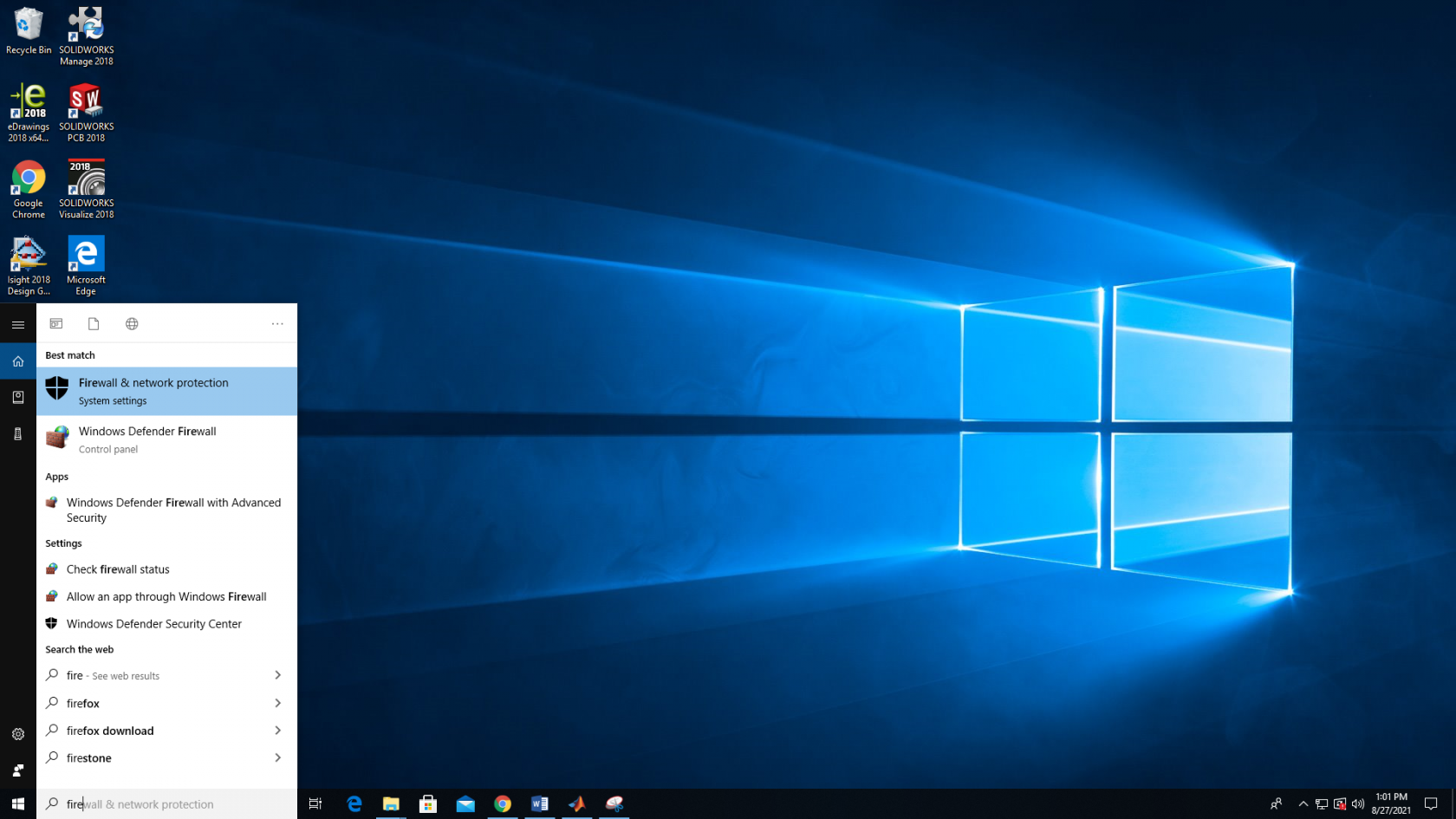
1. Open the start menu and search on "Firewall", then open the "Firewall & Network Protection" program.
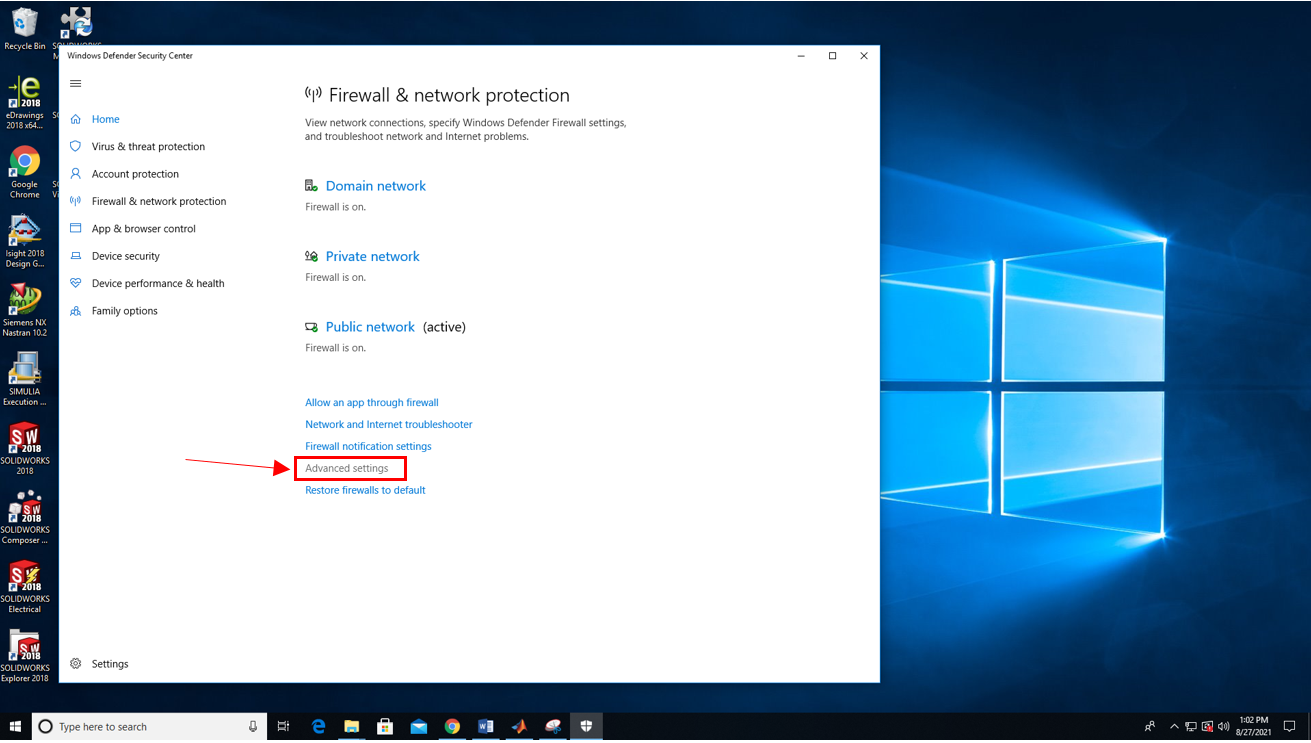
2. Click "Advanced Settings".
3. Give the program administrator priviledges to continue.
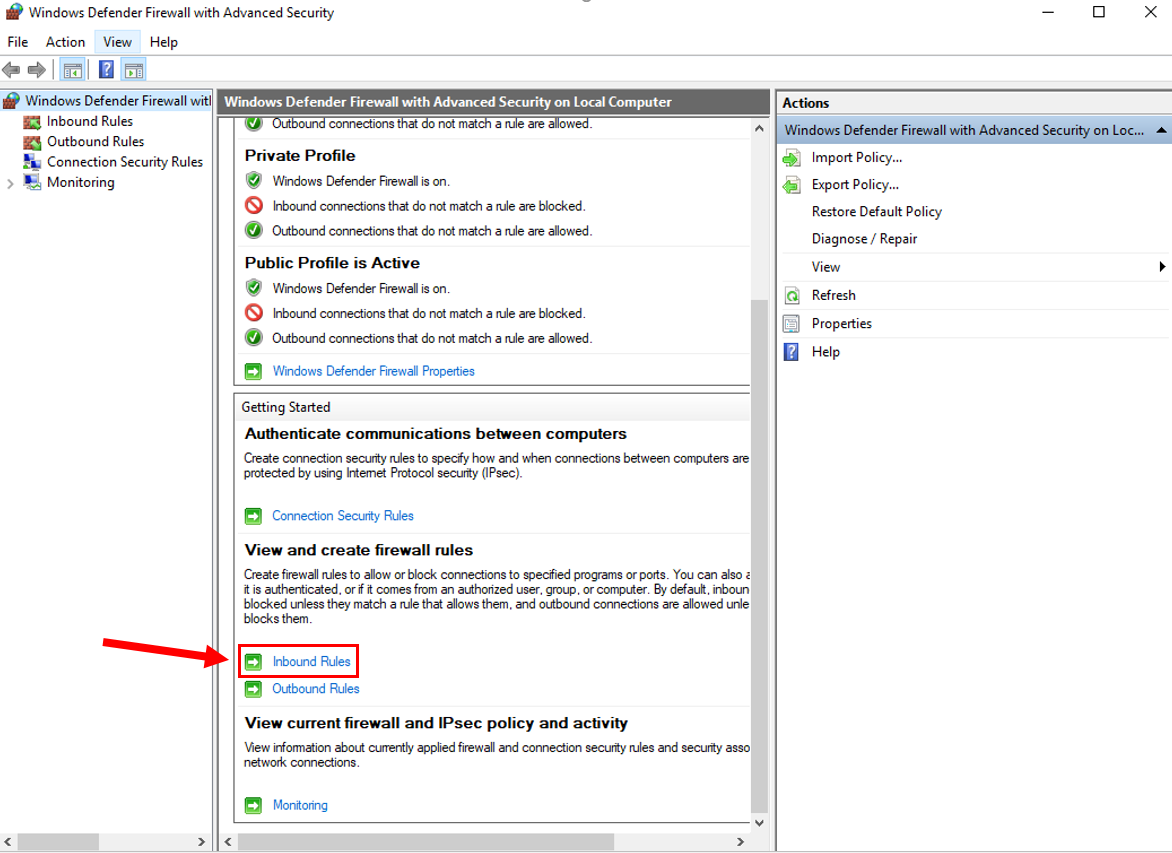
4. Scroll down and click "Inbound Rules".
5. Click "New Rule" on the right hand side.
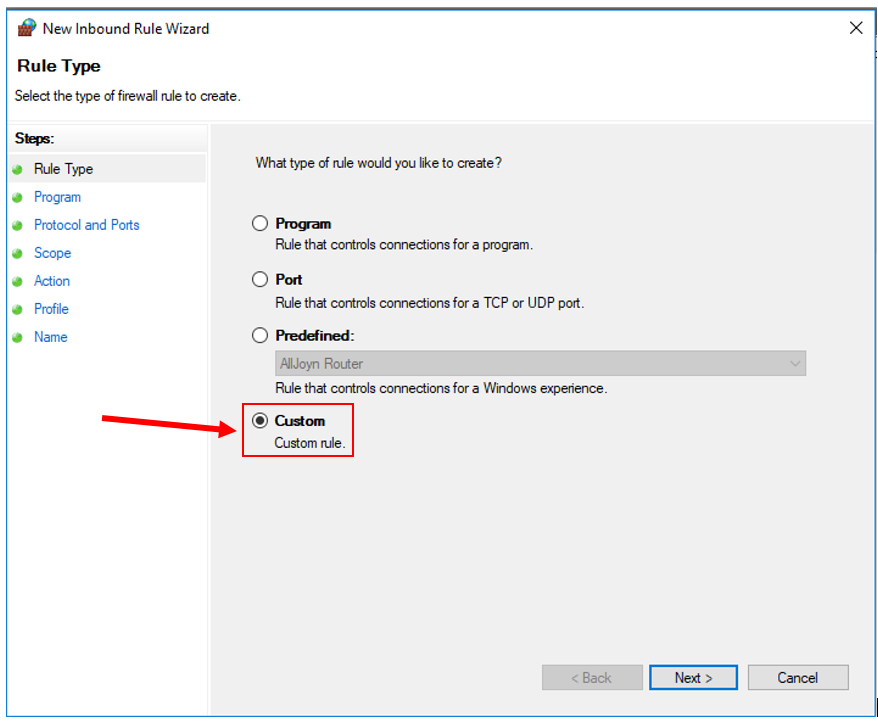
6. Create a Custom Rule.
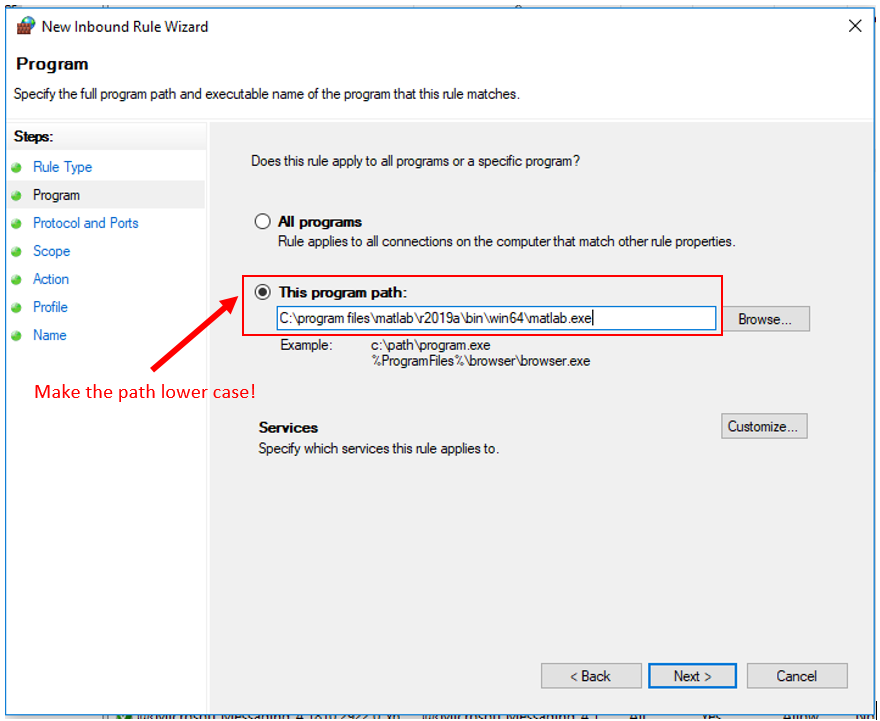
7. Click "Next". Check "This program path", and type in the file path to your Matlab 2022a executable (the one in "\bin\win64\", not just "\bin\" ).
8. Then make the upper-case letters in the path lower-case.
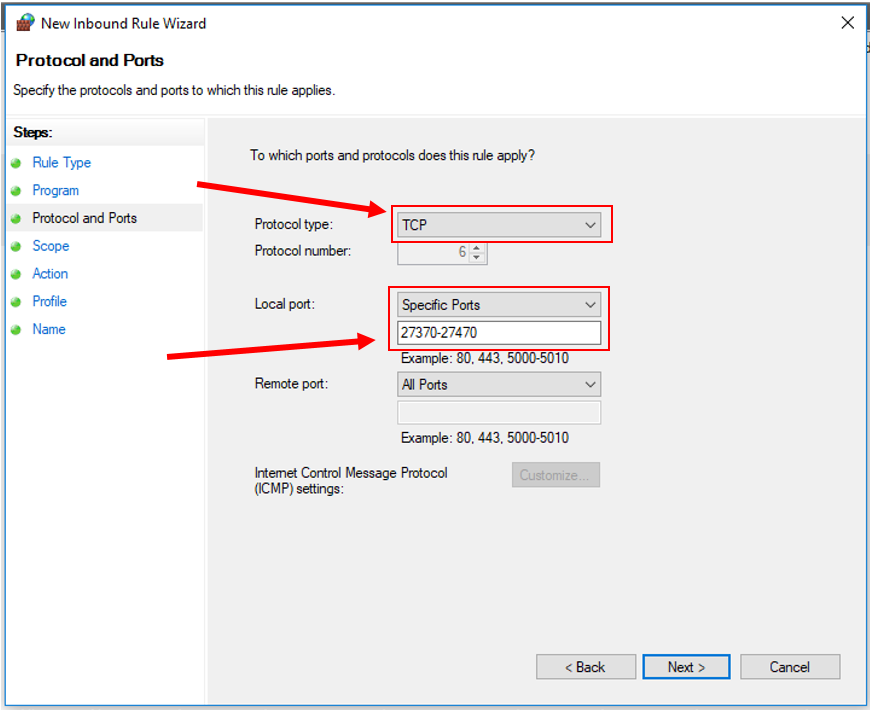
9. Click "Next". Set the Protocol to "TCP" and input the specific port range shown below:
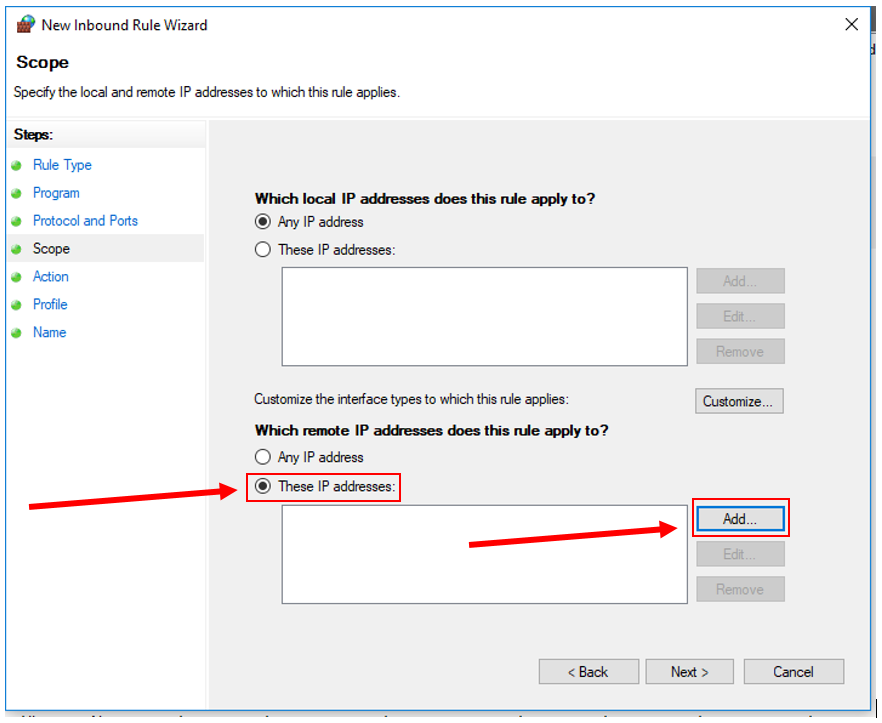
10. Click "Next". Under "Which remote IP addresses does this rule apply to?", check "These IP addresses", then click "Add...".
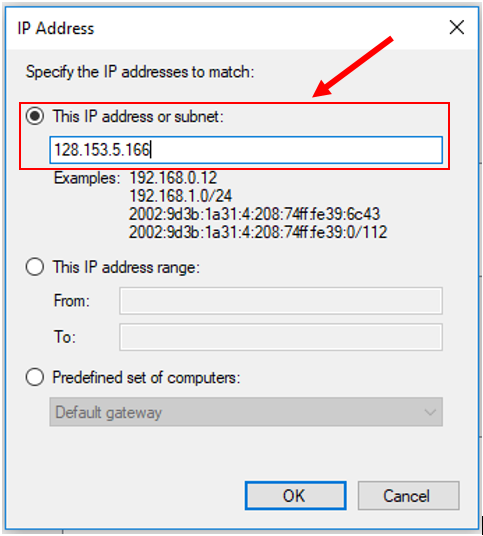
11. Add ACRES head node IP address (128.153.5.166) as shown below. Then click "OK".
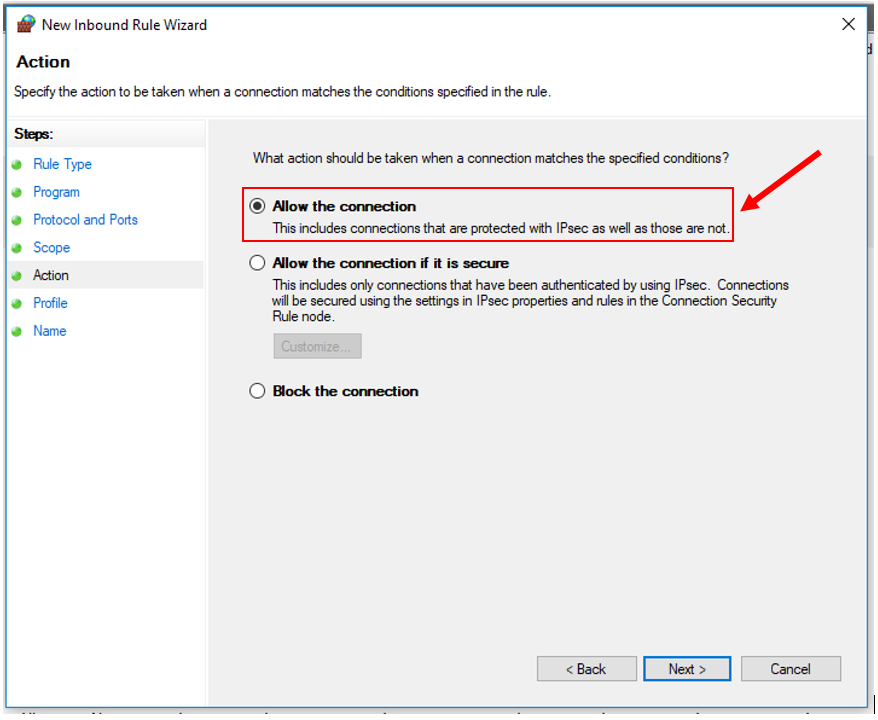
12. Click "Next". Set "Allow the connection".
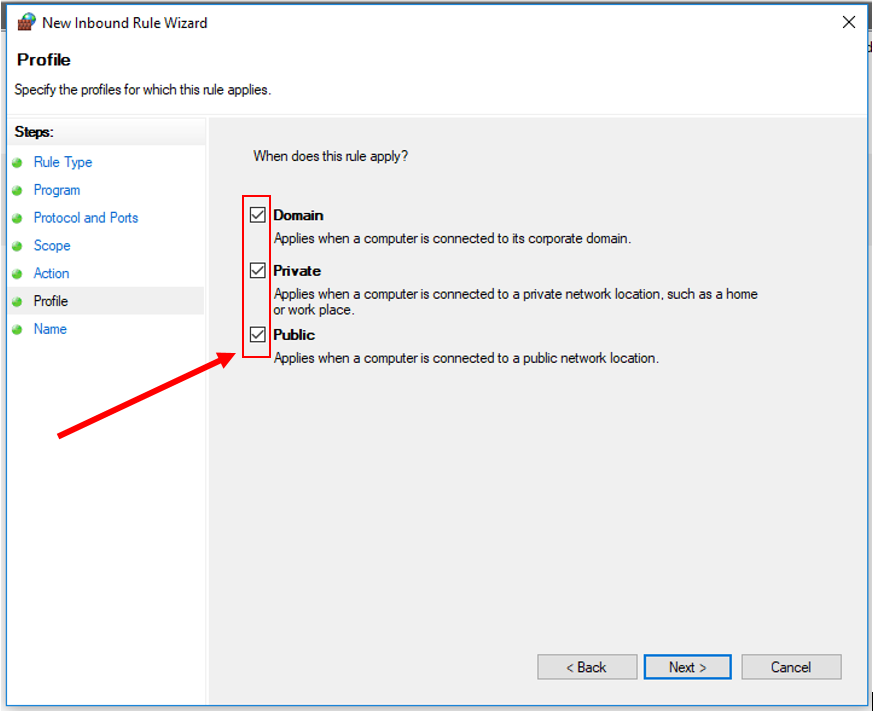
13. Click "Next". Check all three profiles: Domain, Private, and Public.
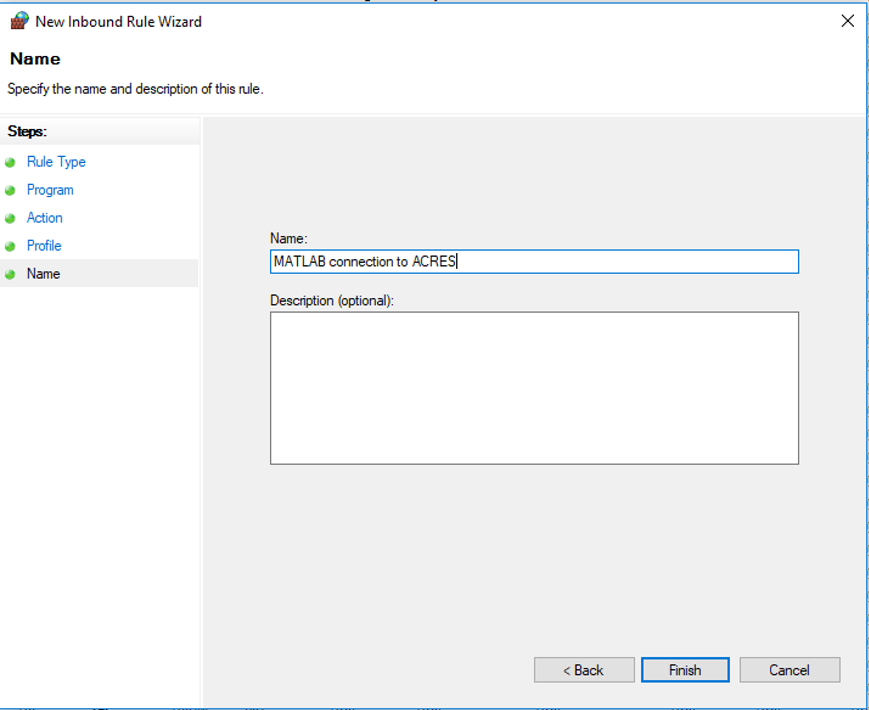
14. Click "Next". Set the name for this rule to something like: "MATLAB connection to ACRES". Add a description, if you'd like. Then click "Finish".
15. You should see the new rule, with the name you gave it, enabled in your list of rules. You may now close the firewall configuration windows.
Linux RedHat / CentOS (FirewallD)
If your Linux machine uses the firewall software "FirewallD" (check by running "which firewalld"), execute the following commands in a terminal:
sudo firewall-cmd --new-zone=ACRES --permanent
sudo firewall-cmd --reload
sudo firewall-cmd --zone=ACRES --add-source=128.153.5.166 --permanent
sudo firewall-cmd --zone=ACRES --add-port=27370-27470/tcp --permanent
sudo firewall-cmd --reload
Linux RedHat / CentOS (iptables)
If you are not using FirewallD, you can likely use "iptables". (Check if you have iptables by running "which iptables", but first check if you are using FirewallD, because that runs on top of iptables, and if you use iptables commands to change settings, FirewallD will not know about the changes)
If you are not using FirewallD, and are only using iptables, Execute the following commands in a terminal:
sudo iptables -A INPUT -p tcp -s 128.153.5.166 --match multiport --dports 27370:27470 -j ACCEPT
sudo iptables-save > /etc/sysconfig/iptables
Step 2. Download Integration Scripts
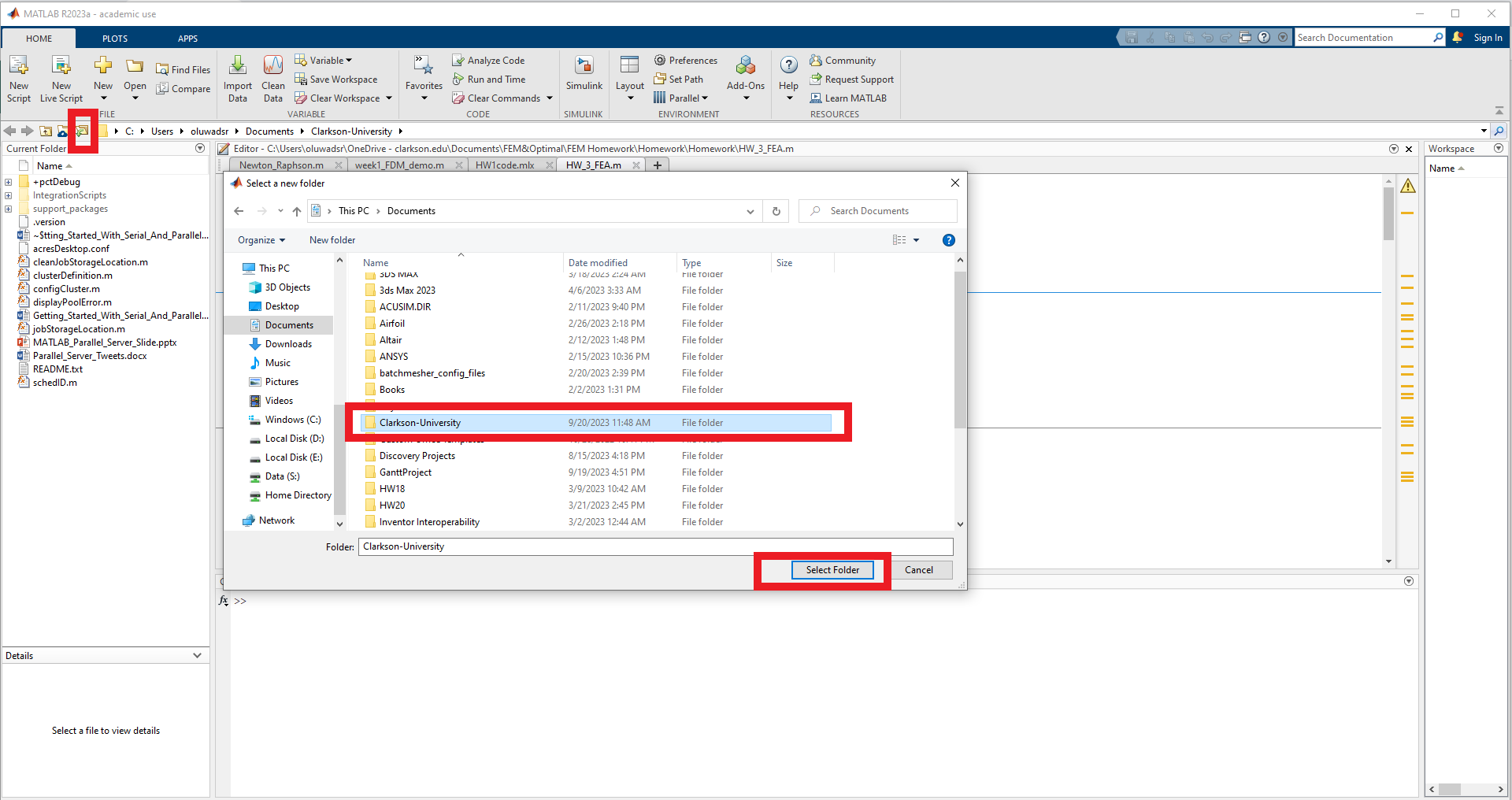
Download the following example integration: Clarkson-University.zip , and unzip it to a local directory.
You may have to right-click on the link above and then "Save link as ..." and select the location to save the file on your computer.
Step 3: Configure the ACRES Cluster Profile and Validate it
Unzip the Cluster Configuration Scripts
Open MATLAB, on MATLAB main window, navigate to the folder where you unzipped and saved the configuration files
In the MATLAB command window type, "configCluster" only needs to be called once per version of MATLAB.
This should prompt you to enter your username on acres.
Cluster Profile Configuration and Validation
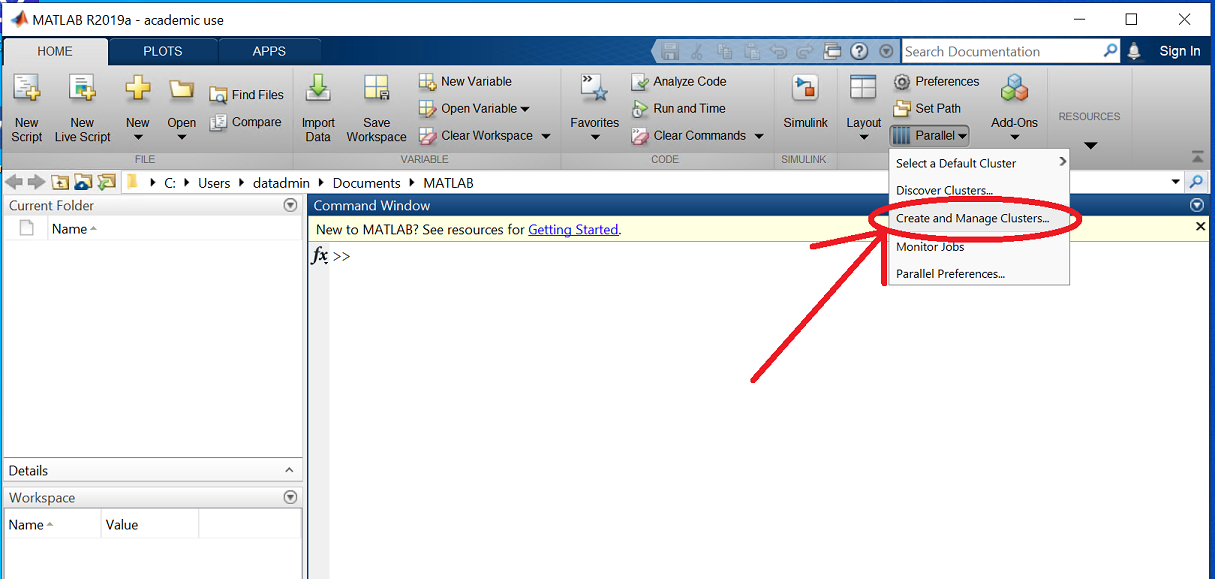
Open MATLAB and click the 'Parallel' drop-down menu item, then click on 'parallel > Create and Manage Clusters...':
This should have opened a new window ('Cluster Profile Manager').
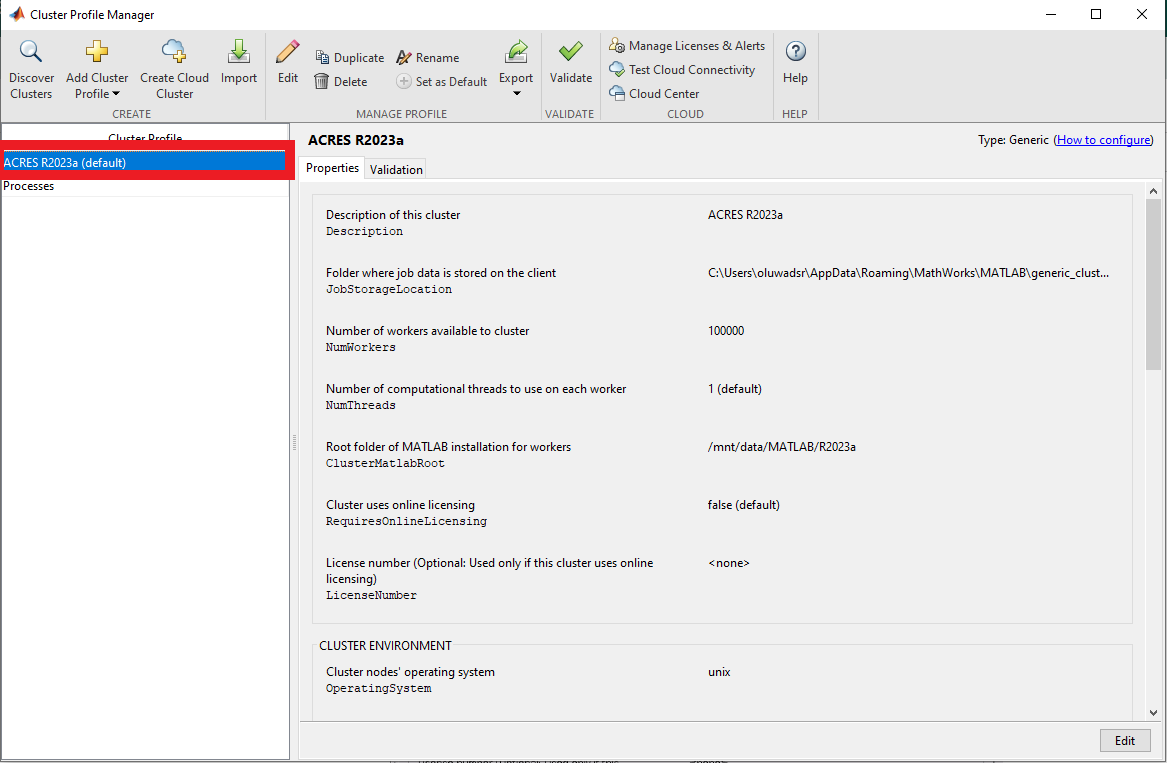
The new profile, ACRES R2023a should be shown and would have been made default.
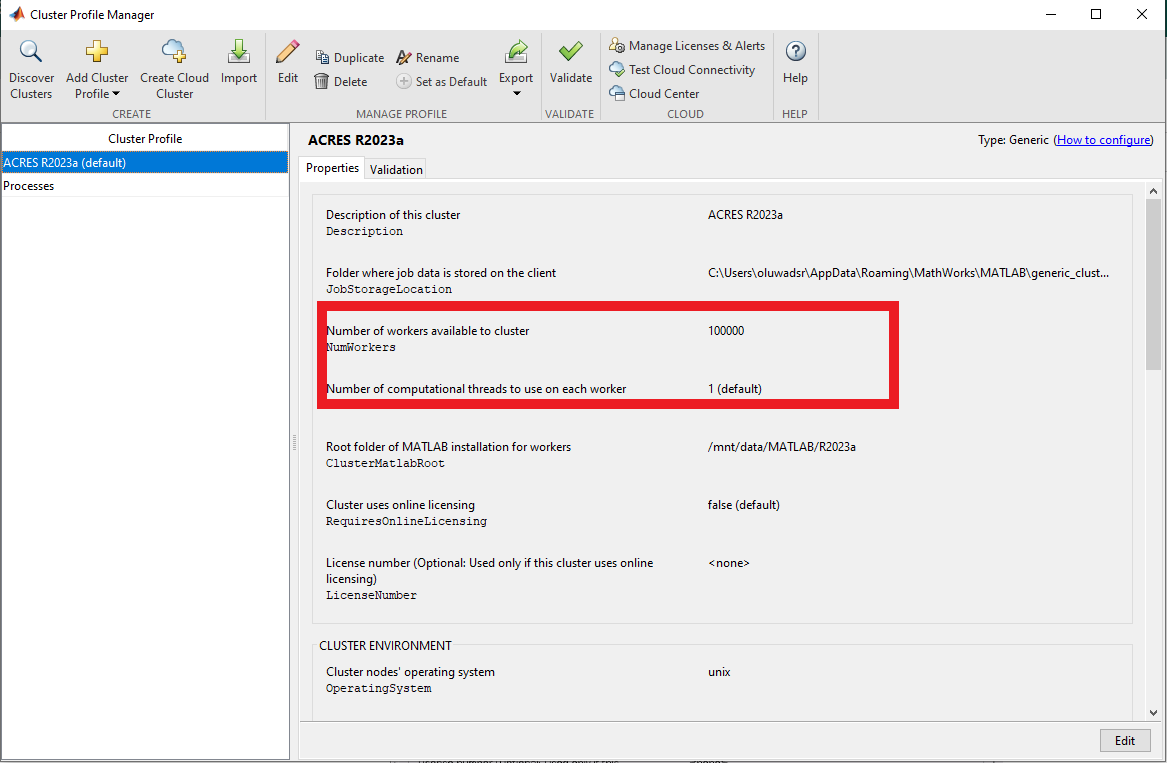
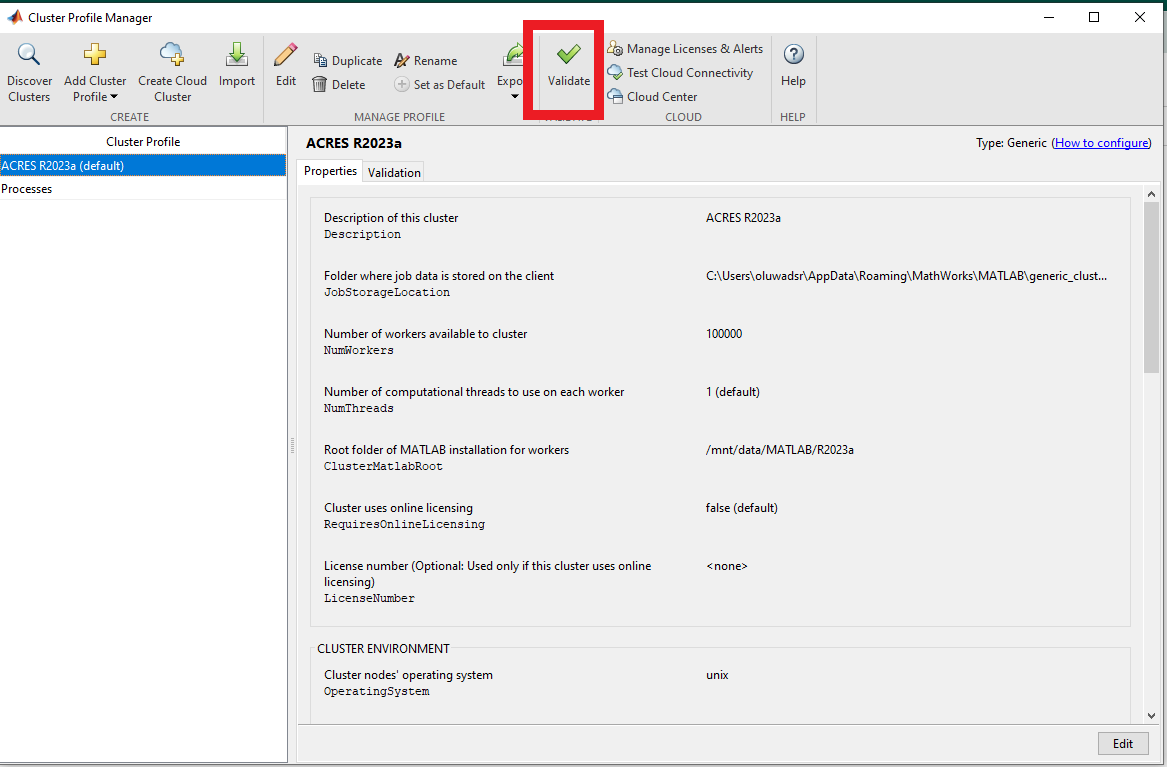
After setting the configuration profile, you will see that, by default, the number of workers is set to 100000, and the number of threads per worker is set to 1. You can specify the number of workers when you submit a job . Note that we have two threads per CPU on the compute nodes, so you can change the number of threads to two.
To run with a different number of tasks/nodes, and threads click "edit", and change the number of workers value as desired.
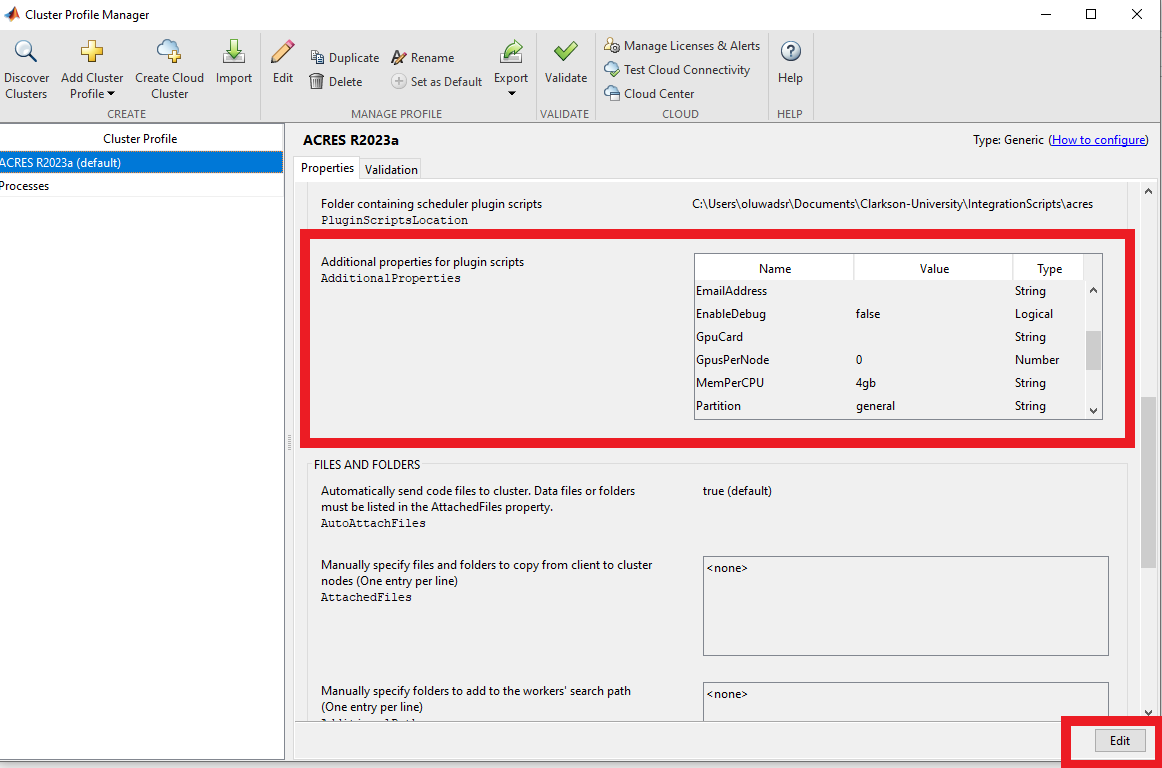
You may also set the ACRES partition in the "partition" field below that. You may use the "dev" partition for running a quick test (e.g. the Validation in the next section), but for running intensive jobs, please use 'general' or another appropriate partition. Also, you can put your email in the email address field.
Click "Done" to save the changes.
Test Configuration
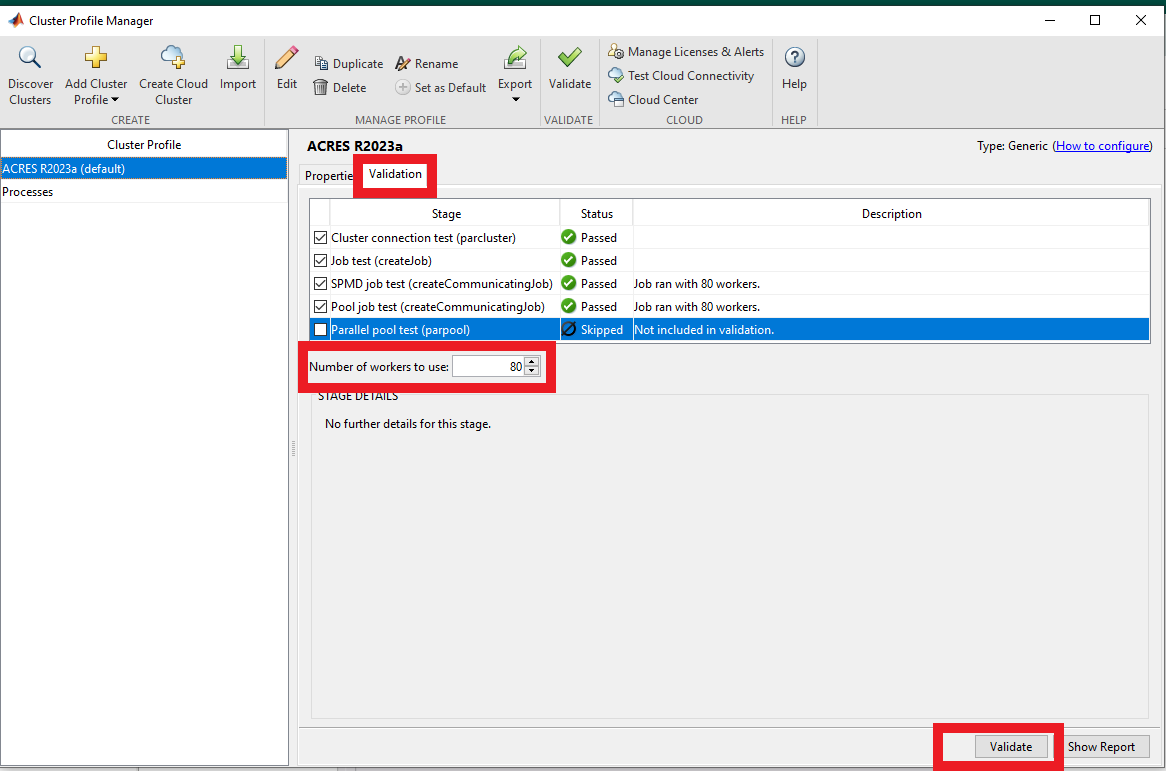
Now select the 'ACRES R2023a' profile and set the appropriate number of workers for your partition. (Think of a "worker" as a "core" in this case) for instance, 80 workers will run on 2 nodes. If you skipped "Step 2", and only intend to run batch jobs, uncheck the last validation test ("parpool").
Go to validation, increase the workers to decent values, like 80 or as desired.

Dev Partition
You may use the "dev" partition with up to 80 workers (there are 2 nodes in the dev partition) for running the Validation. (Edit the appropriate field in "Additional Properties" in the Properties tab to set the partition)
Click the 'Validate' button:

The validation process (and every attempt to run a job on ACRES from here on) will ask you for your credentials to the cluster. You may use username/password authentication, or generate an SSH key pair and use that if you prefer. The validation tests may take some moments to run. Make sure to select a number of nodes to run the validation against that will allow submission of the job to your selected partition.
If you were going through the "Method 1: Run in Batch Mode on ACRES" instructions, continue at Step 4.
Errors?
If one of the Validation stages fails, click on that stage to view the error output. Then check if the error corresponds with one listed in the Troubleshooting section on this page, and try the solutions given there.
For Subsequent Jobs
To run subsequent jobs after the first-time set-up, simply perform Step 2 after starting a new Matlab session. Then you should be able to run any Matlab Parallel commands, using ACRES resources.
Troubleshooting
If MATLAB is unable to start a parallel pool (parpool) (i.e. the fifth validation stage fails):
"No Route to Host" Error
If you get an error that says "No Rout to Host", this is likely a firewall problem, check your firewall settings that you set in Step 1.
"UnknownHostException" Error
If you get an "UnknownHostException" error, then you probably need to repeat Step 2, using "pctconfig" to set the hostname. Be sure that you do this first thing every time you start a new Matlab session and want to compute in parallel on ACRES.
Running an Interactive Parallel Session
If all Validation stages passed, you should be able to run interactive parallel jobs on the cluster. This includes running parallel scripts by simply pressing the green "Run" button in the script editor, or running parallel commands from the command line.
Submitting a Batch Job to the Cluster and Retrieving Results
You may submit a batch job to the cluster by executing the following Matlab commands:
%create a cluster object based on the 'ACRES' cluster profile.
c=parcluster('ACRES')
%after creating the cluster object you can set other properties of it, such as the partition. These override whatever is set in the "cluster profile". Look at the cluster profile configuration menu to see what other variables you may set.
c.AdditionalProperties.partition='general'
%set the number of workers that you'd like to run your job with (NOTE: Matlab will allocate one more core than the number of workers you select here, because one core is used as a coordinator for the others that actually run your job.)
n=79
%submit a job to run a script called 'script_name' (which should be located in the current MATLAB working directory), and store the job object in "myjob".
myjob=c.batch('script_name','Pool',n)
%alternatively, you may also run a function, with specific outputs, instead of a script. Do this via the following syntax:
myjob=c.batch('function_name', Noutputs, {cell, array, of, inputs},'Pool',n)
%where Noutputs is the number of outputs to expect from the function, and the next argument is a cell array of the function inputs.
%Take note of the Job ID by executing:
Jobid = myjob.ID
Notice
You will need to know the Job ID to access the results of your Job.
Once your batch job is submitted, you may close Matlab, and retrieve the results later when the job is finished. To access the results, when the job is finished, you may either use Matlab commands, or the Matlab Job Monitor.
Retrieving Results Through the Matlab command window:
Open Matlab.
Change the Matlab working directory to the folder from which you ran your job. You need to do this before running the following commands:
%create a cluster object based on the 'ACRES' cluster profile.
c=parcluster('ACRES')
%find the job, based on the job id. (replace the "1" with whatever your job ID is, below)
myjob = findJob(c,'ID',1)
%now load the job. If you ran it as a script, use the following, and the variables at that existed at the conclusion of your job will be loaded into your current workspace.
load(myjob)
%if you originally ran the job as a function, use the following to fetch the outputs
outputs = fetchOutputs(myjob)
Retrieving Results Through the Matlab Job Monitor
Important!
To retrieve the results through the Matlab Job Monitor, the Job Monitor window needs to be opened for the first time in the session while the Matlab working directory is the same directory that you ran the job from. (Note that if you close Matlab with the Job Monitor Open, it will re-open automatically on start-up, so you will need to make sure the working directory is correct on start-up, in that case)
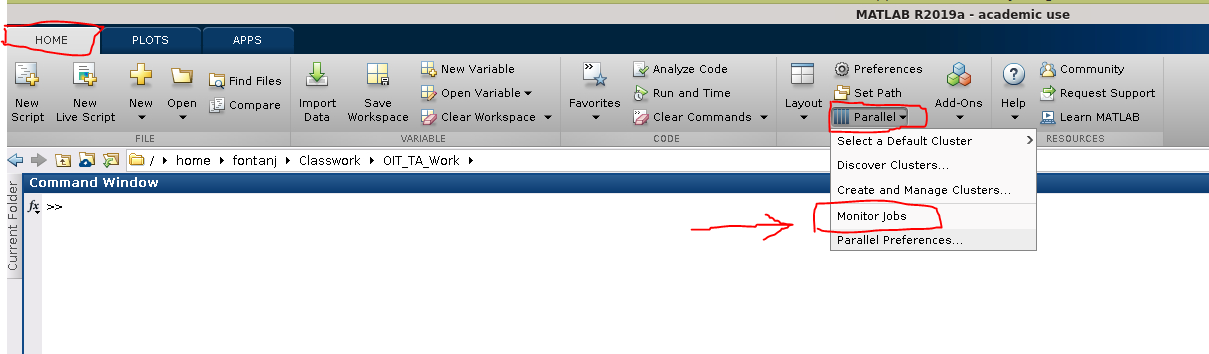
To retrieve the results through the Matlab Job Monitor, the Job Monitor window needs to be opened for the first time in the session while the Matlab working directory is the same directory that you ran the job from. This means you must change the Matlab working directory to your job directory before clicking on -> "parallel" → "Monitor Jobs", as shown below:

Note that if the Job Monitor was open the last time that you closed Matlab, it will again re-open automatically on startup. This means that, in order to make sure the Matlab working directory is your job directory when the Job Monitor opens, you will need to force the Matlab working directory to be your job directory on startup. One way to do this is to start Matlab from the command line in your job directory. Or you may also circumvent the problem by simply starting Matlab, closing the Job Monitor, then closing and re-opening Matlab, changing the working directory to your job directory, then finally re-opening the Job Monitor.
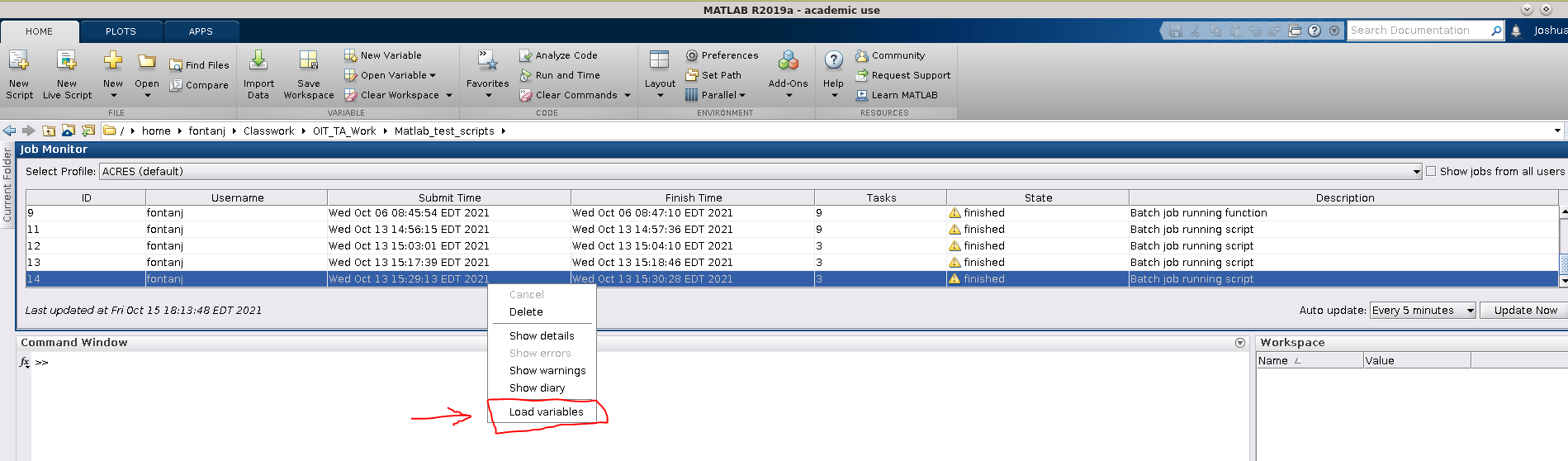
Once the Job monitor is open, and has detected your job, you may right-click on the job listing to load the job's variables, show any warnings or errors, etc.
Viewing Job Console Output
To view the Job console output, navigate to the folder from which you started the job. Then look into the folder for your particular job. (i.e. if your Job had ID=2, look in the "Job2" folder).
The output from your job should be in the "Task1.diary.txt" file inside that folder.
Notice
You may notice that the "*.diary.txt" files from the other Tasks are empty, but this is OK. It does NOT mean that only one Task of your job actually ran. Tests have shown the performance of a job is correlated with the number of Tasks, even though only Task1 produces diary output.
Custom queue submission parameters
To submit jobs with customized parameters, you must edit one of the scripts included in the template configuration. For a communicating job, edit "communicatingSubmitFcn.m"; for a non-communicating job "independentSubmitFcn.m". In either file, search for "additionalSubmitArgs =", and add your customizations there.